Co-registration of BOLD activation area on
Co-registration of BOLD activation area on3D brain image (Courtesy Siemens)
For co-registration single subject fMRI data are commonly overlaid on MR anatomic images acquired during the same session. The anatomic reference is typically from a 3D MR sequence (e.g., MP-RAGE) acquired with high-resolution voxels (e.g., 1mm x 1mm x 1mm). The voxels should be isotropic (perfect cubes, equal in each dimension) to allow the data to be rotated, re-sliced, and manipulated.
Many of the same image correction and alignment techniques for data pre-processing are also used to perform co-registration. These are more fully described in the reference by Klein et al (2009) and the textbook chapters by Ashburner and Friston below. Nearly all of these methods begin with image resampling using interpolation techniques followed by rigid body transformations (translations, rotations, zooms, and shears). An optimization protocol is then employed, typically by minimizing a cost function which measures the degree of disparity between the fMRI and anatomic data after each iteration.
 Image from Talaraich Atlas (1988)
Image from Talaraich Atlas (1988)
Advanced Discussion (show/hide)»
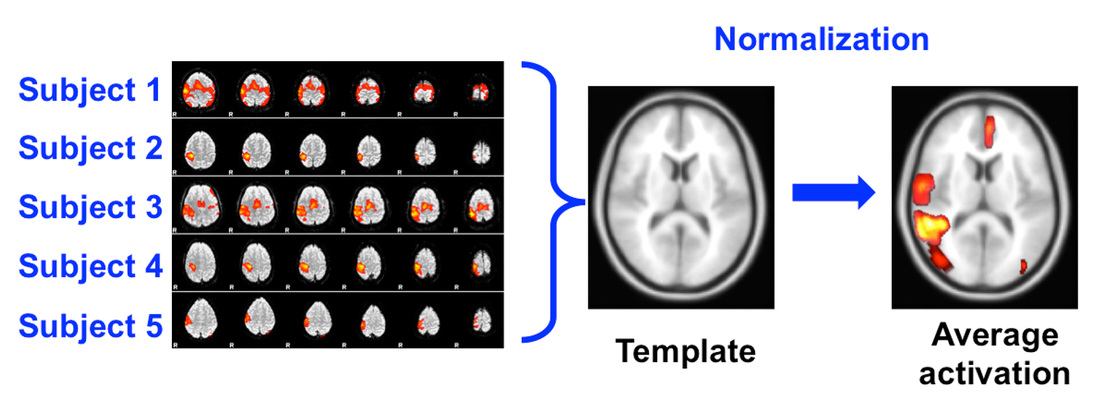
Image Corrections Applied to Anatomic Images in Co-registration
Just as the raw fMRI data can be distorted, so can the T1- or T2-anatomic images used for co-registration. These anatomic images typically must also undergo pre-acquisition or pre-processing corrections steps. Examples include:
1. Gradient non-linearity compensation. A method such as "Gradwarp" is commonly used to correct for image distortion secondary to gradient non-linearities. This is done as part of routine imaging processing by many commercial scanners automatically.
2. B1-non-uniformity correction. This refers to signal intensity differences caused due to variations in the B1 transmit field or RF-signal reception by a non-uniform head coil. Calibration scans may be performed to account for these spatially dependent changes in signal during the image acquisition phase.
3. Dielectric effect non-uniformity correction. This refers to signal intensity variations due to standing waves and dielectric effects that become increasingly problematic at 3T and above. These may be reduced by intensity correction filters or histogram peak-sharpening algorithms such as the N3 technique.
Considerations for Multimodality Co-registration
If the anatomic MR images have been acquired during the same session as the fMRI study, then header information in the data will allow fully automated initial alignment to be performed. If the anatomic data comes from a different modality such as CT, or if the patient has moved significantly between the functional and anatomic MR sequences, a manual adjustment may be required to make sure that the image sets are in gross correspondence. In some cases initial alignment between data sets is performed by manual matching of specified points on the images. These points may be an easily recognized anatomic structure (such as the midline anterior commissure) or an external landmark (such as a fiducial marker attached to the scalp).
The Talairach Patient
Virtually nothing is known about the woman whose brain was used for the Talairach atlas. Anectdotal published references from the 1990's suggest that she may have been French, 60-69 years old, right-handed, and possibly an alcoholic. I have tried trace down these statements from various authors, and to date have been unable to verify any of their demographic statements. The T&T Atlas of 1988 says nothing except the subject was female, and unfortunately both Talariach and Tournoux are no longer alive to comment further. I suspect that because T&T worked in Paris the woman was merely assumed to be French, and the reference to alcoholism was likely just a slur on the French who do love their wine. I also question that she was in her 60's, as her sulci and ventricles seem rather small to me for an average person in this age group. If any readers have direct knowledge about this subject, please let me know!
Ashburner J, Friston KJ. Rigid body registration. In: Frackowiak RSJ (ed). Human Brain Function, 2nd ed. London: Academic Press, 2004. (pdf made available on-line by the authors at http://www.fil.ion.ucl.ac.uk/spm/doc/books/hbf2/)
Ashburner J, Friston KJ. Spatial normalization using basis functions. In: Frackowiak RSJ (ed). Human Brain Function, 2nd ed. London: Academic Press, 2004. (pdf made available on-line by the authors at http://www.fil.ion.ucl.ac.uk/spm/doc/books/hbf2/)
Ashburner J, Friston KJ. High-dimensional image warping. In: Frackowiak RSJ (ed). Human Brain Function, 2nd ed. London: Academic Press, 2004. (pdf made available on-line by the authors at http://www.fil.ion.ucl.ac.uk/spm/doc/books/hbf2/)
Brett M, Christoff K, Cusack R, Lancaster JL. Using the Talairach atlas with the MNI template. Medical Research Council, 1997. (Downloaded from brainmap.org 5/20/16)
Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. NeuroImage 2009; 48:63-73.
Klein A, Andersson J, Ardekani BA, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage 2009; 46:786-802. (Good review of various functional-structural co-registration and spatial normalization algorithms)
Lancaster JL, Woldorff MG, Parsons LM, et al. Automated Talairach atlas labels for functional brain mapping. Hum Brain Mapp 2000; 10:120-131.
Talairach J, Tournoux P. Co-planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System - an Approach to Cerebral Imaging. New York: Thieme Medical Publishers,1988.
What are the steps for preprocessing fMRI data?
How are those activation "blobs" on an fMRI image created, and what exactly do they represent?